Graduate student Sean Fletcher has received acceptance of his paper to the International Symposium on Visual Computing (ISVC 2025). The paper, co-authored with Dr. Yuqi Song, Dr. Xin Zhang, and in collaboration with the Biology Department under Dr. Doug Currie, marks a major achievement in interdisciplinary research at USM. The project demonstrates the power of interdisciplinary research, combining expertise in computer vision, machine learning, and cell biology to advance biomedical imaging tools.
The study, “Interpretable Tile-Based Classification of Paclitaxel Exposure”, develops a three-phase tiling-and-aggregation pipeline to classify subtle drug-induced changes in cell morphology. Conventional full-image deep learning models often dilute local information, making it difficult to detect fine-grained differences across treatment conditions. To address this, the researchers partitioned each high-resolution phase-contrast microscopy image (1600×1200 pixels) into smaller, non-overlapping tiles. These tiles were resized to 224×224 pixels and processed by a ResNet-50 convolutional neural network pretrained on ImageNet but adapted for grayscale biomedical images. Tile-level features were evaluated using both a fully connected head and a k-nearest neighbors (k-NN) classifier in embedding space.
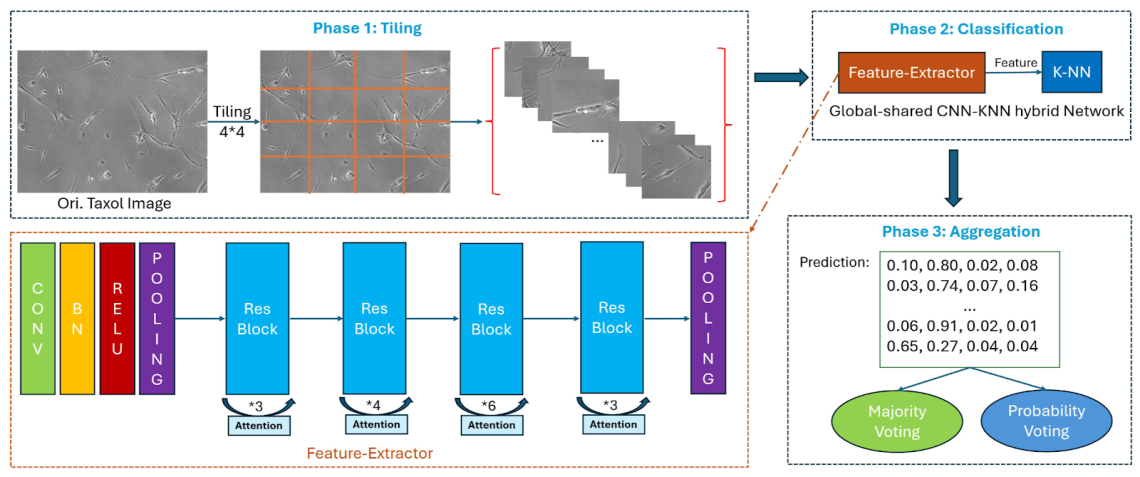
Once each tile was classified, the predictions were aggregated at the image level using either majority voting or probability-weighted voting, which stabilizes decisions by averaging across tiles. This ensemble-style process proved highly effective: the pipeline achieved 97% accuracy, outperforming the previously published CNN+kNN baseline by 20 percentage points. The researchers also systematically compared different tiling grid sizes (from coarse 2×2 splits to fine 12×12 splits) and found that performance peaked at a 6×7 grid, which balances local detail with sufficient spatial context.
Beyond raw performance, the team conducted interpretability analyses using Grad-CAM and Score-CAM heatmaps. These visualizations revealed that the model often focused on peri-cellular textures and micro-environmental cues, in addition to direct cellular morphology. While this explains the performance boost, it also underscores the challenges of robustness when applying AI models across laboratories with different imaging conditions. The authors recommend future work to align model attention more closely with biologically meaningful cell structures, ensuring better generalization.

